Research Focus /目的
Our lab investigates the fascinating mechanisms that coordinate the cell cycle with developmental and physiological processes in multicellular organisms. Disruptions to this coordination can lead to serious health consequences, including cancer, tissue degeneration, and ageing. To better understand this critical coordination, we focus on a group of evolutionarily conserved proteins called “Cell Cycle Regulators (CCRs).” Recent research, including our own, has revealed that CCRs not only control the cell cycle but also regulate cell fate and differentiation through previously unknown molecular functions, making them “intracellular proliferation-differentiation coordinators” (Kimata et al., 2020).
我们的实验室研究多细胞生物中协调细胞周期与发育和生理过程的迷人机制。这种协调的破坏可能导致严重的健康后果,包括癌症、组织退化和衰老。为了更好地理解这一关键协调,我们专注于一组被称为“细胞周期调控因子(CCR)”的进化保守蛋白。最近的研究,包括我们自己的研究,揭示了CCR不仅控制细胞周期,还通过以前未知的分子功能调控细胞命运和分化,使它们成为“细胞内增殖-分化协调因子”(Kimata et al., 2020)。
To explore these novel functions, we use advanced genetics and cutting-edge technologies such as in vivo imaging, multi-omics analysis, and artisan biochemical techniques in the fruit fly Drosophila melanogaster, a powerful model organism for multicellular research. We also examine these mechanisms in human cells and mammalian organoids to translate our findings from Drosophila to humans. Our research aims to decipher the evolutionarily conserved molecular mechanisms that integrate cell proliferation with the development and physiology of multicellular organisms. Ultimately, our findings will contribute to the development of new approaches to treat incurable diseases such as cancer, while bridging the gap between cell biology and developmental biology.
为了探索这些新功能,我们使用先进的遗传学和尖端技术,如果蝇Drosophila melanogaster体内成像、多组学分析和工匠生化技术,这是多细胞研究的有力模型生物。我们还检查这些机制在人类细胞和哺乳动物类器官体中的表现,以将我们在果蝇中的发现转化为人类。我们的研究旨在破译协调多细胞生物的细胞增殖与发育和生理的进化保守分子机制。最终,我们的发现将有助于开发治疗癌症等无法治愈的疾病的新方法,同时弥合细胞生物学和发育生物学之间的鸿沟。
We welcome potential collaborators and students who are interested in contributing to our ongoing projects, which include:
我们欢迎有兴趣为我们正在进行的项目做出贡献的潜在合作者和学生,项目包括:
1. Unveiling novel cell cycle-independent functions of CCRs in cell fate choice and differentiation control / CCR在细胞命运选择和分化控制中的新型非细胞周期依赖功能
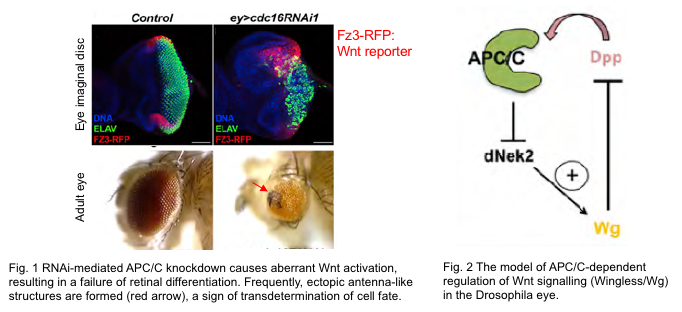
In vivo, various cell behaviors such as cell differentiation and morphogenesis are orchestrated by intra/intercellular signal transduction and transcription factors mainly through gene regulation. Recent research indicates that some CCRs directly regulate the activities of these developmental factors in concert with cell cycle regulation. One of the prime examples is the Anaphase-Promoting Complex (APC/C), a master CCR, which inhibits the transcriptional activity of a major extracellular signaling pathway, Wnt pathway, by degrading its positive modulator Nek2 in the Drosophila eye imaginal disc. Through this mechanism, APC/C coordinates the cell cycle exit of progenitor cells with the induction of differentiation into specialized cell types (Martins et al., 2017).
在多细胞生物身体内,各种细胞行为,例如细胞分化和形态发生,主要通过基因调控由细胞内/细胞间信号传导和转录因子协调。最近的研究表明,一些CCR直接调控这些发育因子的活性,与细胞周期调控协同作用。其中一个主要例子是促间期促进复合体(APC/C),一种主要的CCR,通过降解果蝇眼睛想象盘中Wnt途径的正调节因子Nek2,抑制了主要胞外信号传导途径的转录活性。通过这种机制,APC/C协调了祖细胞的细胞周期退出与分化为专门细胞类型的诱导(Martins et al., 2017)。
We have extended this initial finding by conducting genome-wide screens of CCRs to identify novel non-cell cycle functions of CCRs in Drosophila. Through this screen, we have identified several CCRs that severely impair retinal differentiation upon knockdown. We are currently characterising the underlying molecular mechanisms of some of these CCRs, including CDKs. Furthermore, we are also studying the conserved functions of mammalian orthologs of the multifunctional CCRs in cultured human cells, organoids and the mouse model. This study will enable us to unravel the conserved mechanism of CCRs implicated in human biology and disease.
我们通过对CCR进行全基因组筛选,扩展了这一初步发现,以识别果蝇中CCR的新颖非细胞周期功能。通过这一筛选,我们已经识别出几种CCR,当它们被敲低时严重损害视网膜分化。我们目前正在研究其中一些CCR(包括CDKs)的潜在分子机制。此外,我们还在研究培养的人类细胞、类器官和小鼠模型中多功能CCR的哺乳动物同源基因的保守功能。这项研究将使我们能够揭示与人类生物学和疾病有关的CCR的保守机制。
The genome-wide screen for cell cycle-independent functions of CCRs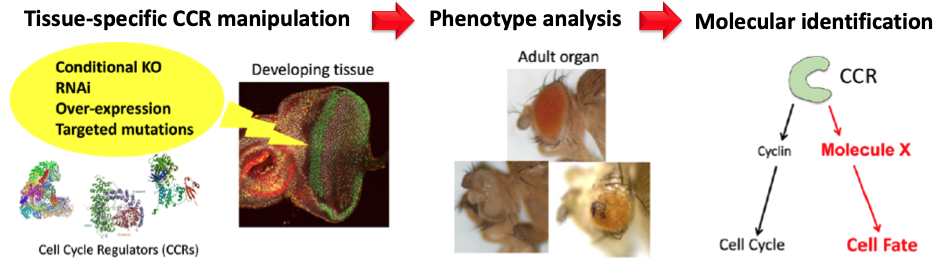
2. Unlocking the Secrets of Dormant Cells: Investigating Regulatory Mechanisms of G0 Phase and Cell Cycle Exit / 休眠细胞的秘密:研究G0期和细胞周期停止的调控机制
A significant proportion of cells in adult animals are in a dormant state and not actively dividing. Tissue-specific stem cells, for example, are often in a reversible quiescent state known as the “G0 phase,” dividing only when necessary to repair tissue damage. Meanwhile, differentiated cells such as neurons and muscle cells have irreversibly exited from the cell cycle and no longer proliferate. The mechanisms that control and maintain these dormant cell cycle states and coordinate them with cell type- and tissue-specific functions remain largely unknown.
成年动物中相当一部分细胞处于休眠状态,并未积极分裂。例如,特异性组织的干细胞通常处于一种可逆的休眠状态,被称为“G0期”,仅在修复组织损伤时才分裂。与此同时,分化的细胞如神经元和肌肉细胞已从细胞周期中不可逆地退出,不再增殖。控制和维持这些休眠细胞周期状态的机制,并将其与细胞类型和组织特异性功能协调,仍然在很大程度上未知。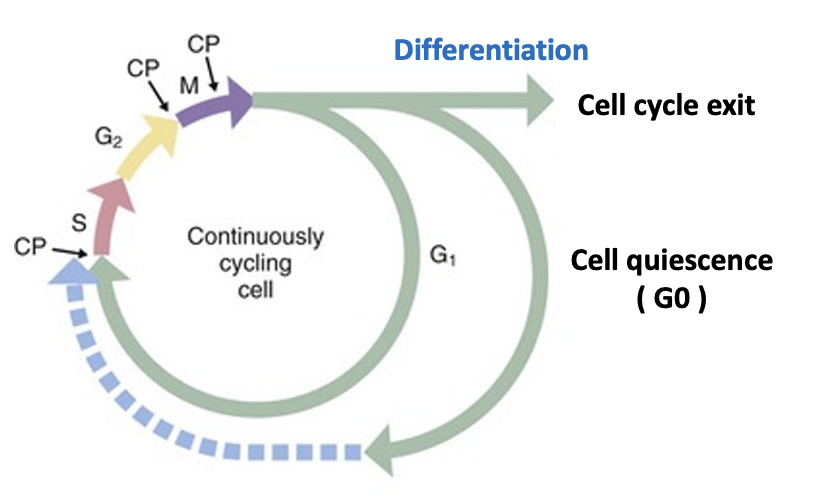
We are investigating these enigmatic cell cycle states both in vivo and in vitro. In Drosophila, we study terminally differentiated tissues such as the brain and gut of adult flies to investigate how the quiescent states of tissue-specific stem cells and differentiated cells are controlled in vivo, and how deregulation of these states affects tissue homeostasis and function. We are also examining the precise regulatory mechanisms that coordinate entry into the G0 phase with genetic and epigenetic states of the genome in cultured human cells, using recently developed rapid protein targeting systems. Our goal is to gain insights into the fundamental mechanisms that control the G0 phase and cell cycle exit, and how their regulation can contribute to tissue homeostasis, tissue degeneration and cancer.
我们正在体内和体外研究这些神秘的细胞周期状态。在果蝇中,我们研究成虫大脑和肠道等终末分化的组织,以调查组织特异性干细胞和分化细胞的休眠状态是如何在体内控制的,以及这些状态的失调是如何影响组织稳态和功能的。我们还在使用最近开发的快速蛋白靶向系统,研究培养的人类细胞中进入G0期与基因组的遗传和表观遗传状态相协调的精确调控机制。我们的目标是深入了解控制G0期和细胞周期退出的基本机制,以及它们的调控如何有助于组织稳态、组织退化和癌症。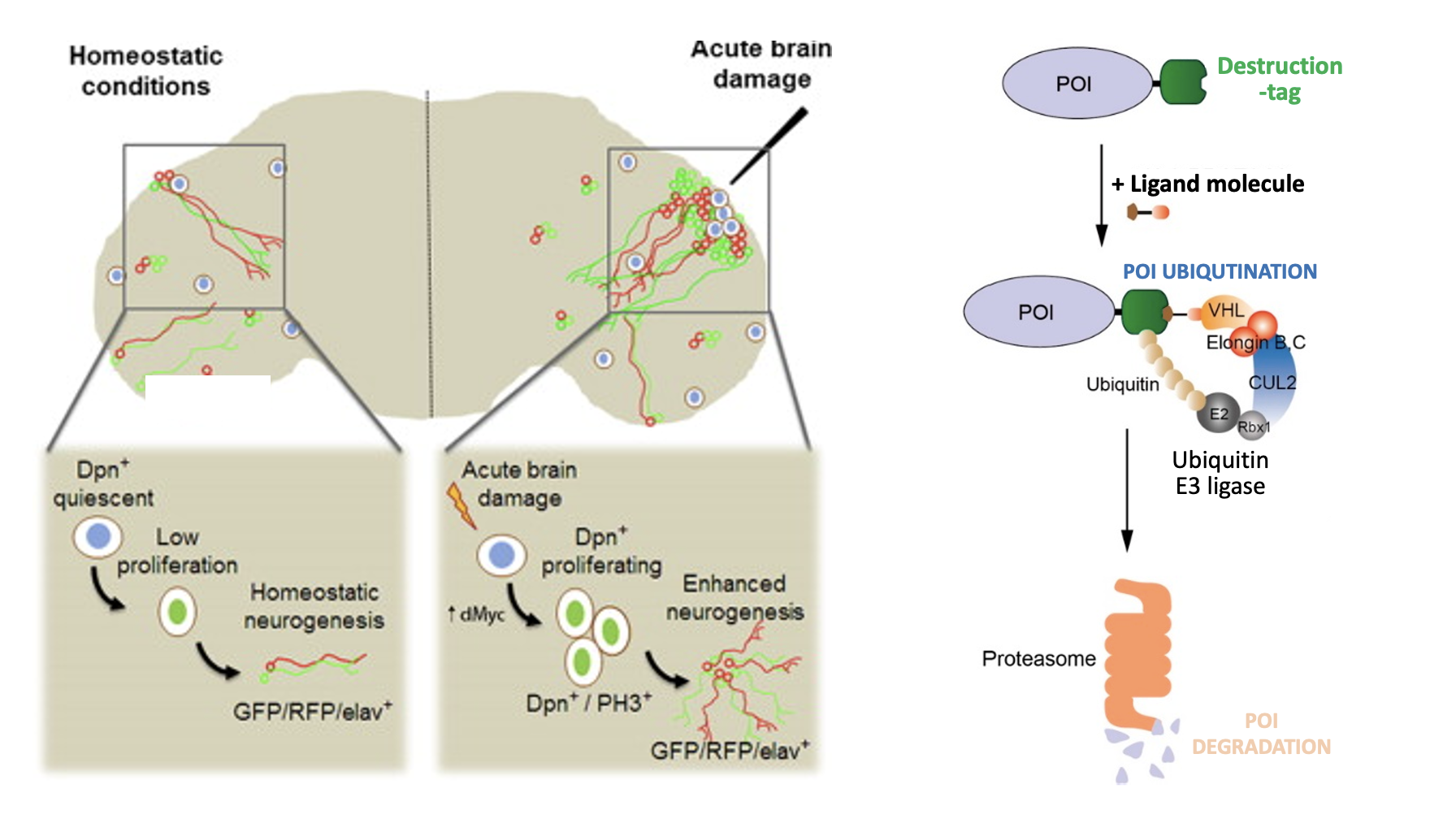
3. Molecular Interactions between CCRs and the Centrosome: Characterisation of Functions in Development and Disease
The centrosome is an organelle found in animal cells that provides spatial and directional control and mechanical strength by regulating the cytoskeleton. In non-dividing cells, centrosomes can transform into primary cilia, which play a critical role in signal transduction and mechanical sensing. Centrosome abnormalities are associated with various disorders, including cancer, macrocephalies and ciliopathies.
中心体是动物细胞中的一种细胞器,通过调节细胞骨架提供空间和方向控制以及机械强度。在非分裂细胞中,中心体可以转化为初级纤毛,对信号传导和机械感应起着关键作用。中心体异常与包括癌症、巨头症和纤毛病在内的多种疾病有关。
There is mounting evidence that various CCRs interact with centrosomes and primary cilia and reciprocally control each other’s functions in multicellular organisms. In collaboration with David Glover’s lab and Pietro Lio’s lab at the University of Cambridge, we previously found that the APC/C activator CDH1 (Fzr in Drosophila) directly binds to a conserved centrosome component, Spd2/CEP192 in Drosophila tissues, through in vivo proteomic analysis. We showed that Spd2 recruits CDH1 onto the centrosome to promote the degradation of Aurora A, a centrosomal substrate of APC/C, which is important for maintaining the number of neural stem cells in the developing adult brain (Meghini et al., 2016). We also found that the protein levels of Spd2 are in turn regulated through APC/C-dependent degradation, and abnormal accumulation of Spd2 affects the function of interphase centrosomes that are crucial for the division axis maintenance of neural stem cells (Meghini et al., 2023). Moreover, in collaboration with Renata Basto at the Institute Curie in France, we demonstrated that this interaction is critical for the centrosome regulation specific to asymmetric division of neural stem cells (Gambarotto et al, 2019).
越来越多的证据表明,不同的CCR与中心体和初级纤毛相互作用,并在多细胞有机体中相互控制各自的功能。与剑桥大学的David Glover实验室和Pietro Lio实验室合作,我们以前通过体内蛋白质组学分析发现APC/C激活剂CDH1(果蝇中的Fzr)直接结合到一种保守的中心体成分Spd2/CEP192(Meghini et al., 2016)。我们展示了Spd2招募CDH1到中心体上促进Aurora A的降解,这对于维持成人大脑发育中神经干细胞的数量非常重要。我们还发现Spd2的蛋白水平反过来通过APC/C依赖的降解进行调控,Spd2的异常积累影响了细胞周期间期中心体的功能,这些对于神经干细胞的分裂轴线维持至关重要(Meghini et al., 2023)。此外,与法国居里研究所的Renata Basto合作,我们证明了这一相互作用对于神经干细胞的非对称分裂中特异的中心体调控至关重要(Gambarotto et al, 2019)。
Furthermore, using the CRISPR genome editing technique to create new dominant mutations in an APC/C cofactor Vihar/Ube2C, we recently showed that APC/C controls the migration of centrosomes to the future oocyte during oocyte specification. This control is mediated by the degradation of Polo/Plk1 kinase, another key centrosomal kinase whose levels are often upregulated in cancer cells (Braun et al., 2021).
此外,通过使用CRISPR基因组编辑技术在APC/C辅助因子Vihar/Ube2C中创建新的显性突变,我们最近展示了APC/C通过降解另一关键中心体激酶Polo/Plk1激酶来控制中心体向未来卵母细胞的迁移,而这种激酶的水平在癌细胞中通常上调(Braun et al., 2021)。
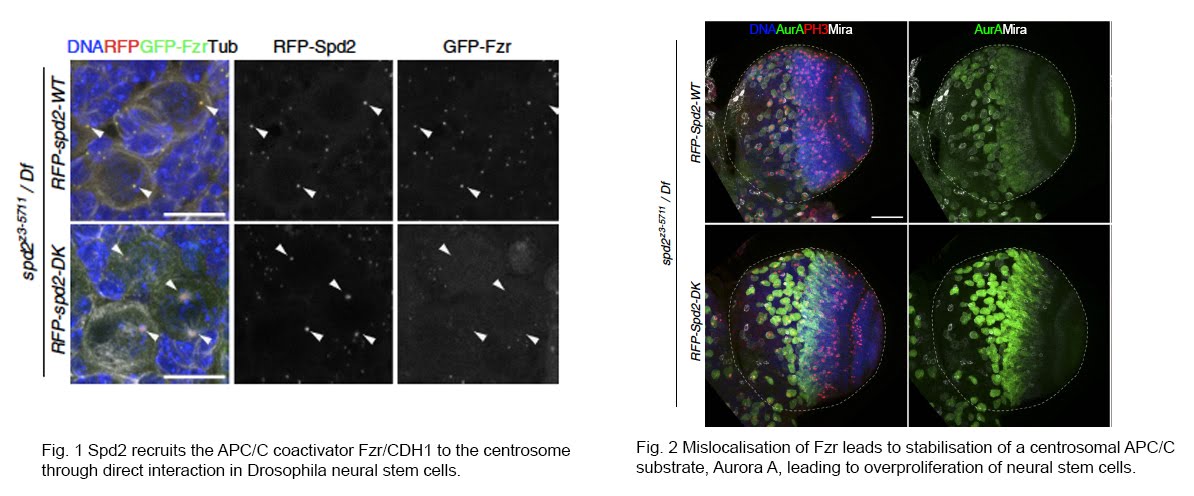
We are continuing to explore the interactions between CCRs and centrosome/cilia components and their functions in various Drosophila tissues and human cells and organoids.
我们将继续探索CCR与中心体/纤毛成分之间的相互作用以及它们在不同的果蝇组织和人类细胞和类器官中的功能。
Recent publications
- Meghini F, Martins T, Zhang Q, et al., APC/C-dependent degradation of Spd2 regulates centrosome asymmetry in Drosophila neural stem cells. EMBO Rep. 2023 April.
- Braun AL, Meghini F, Villa-Fombuena G, et al., The careful control of Polo kinase by APC/C-Ube2C ensures the intercellular transport of germline centrosomes during Drosophila oogenesis. Open Biol. 2021 June.
- Kimata Y, Leturcq M, Aradhya R. Emerging roles of metazoan cell cycle regulators as coordinators of the cell cycle and differentaition. FEBS Letters. 2020 July.
- Ochi T, Quarantotti V, Lin H, et al. CCDC61/VFL3 is a paralog of SAS6 and promotes ciliary functions. Structure. 2020 June.
- Gambarotto D, Pennetrier C, Rynaiawec JM, et al. Plk4 regulates centriole asymmetry and spindle orientation in neural stem cells. Developmental Cell. 2019 Jul.
- Kimata Y. APC/C Ubiquitin Ligase: Coupling Cellular Differentiation to G1/G0 Phase in Multicellular Systems. Trends in Cell Biology. Apr. 2019
- Martins T, Meghini F, Florio F, Kimata Y. The APC/C coordinates retinal differentiation with G1 arrest through the Nek2-dependent modulation of Wingless signalling. Developmental Cell. 2017 Jan.
- Meghini F, Martins T, Tait X (equally contributed), Fujimitsu K, Yamano H, Glover DM, Kimata Y. Targeting of Fzr/Cdh1 for timely activation of the APC/C at the centrosome during mitotic exit. Nature Comms. 2016 Aug 25;7:12607.